Structural Biology Program in Physiology
 All proteins that are integral to the cellular membrane are endowed with the ability to reside and function in a hydrophobic environment. Over the last twenty years, mostly thanks to the dramatic improvement in our ability to express, purify and determine their atomic structure, we have learned a great deal about the principles that govern structure and activity of membrane proteins. For example, the ion channels and transporters that mediate the movement of charged molecular species across the lipid bilayer, are now rather well understood; how G-protein coupled receptors (GPCRs) signal to the inside of the cell upon binding to an extracellular ligand is also being elucidated. Indeed, we have acquired a wealth of knowledge about how membrane proteins are shaped and work to allow solutes and biological signals to traverse and be transmitted across the water-impermeable lipid bilayer.
All proteins that are integral to the cellular membrane are endowed with the ability to reside and function in a hydrophobic environment. Over the last twenty years, mostly thanks to the dramatic improvement in our ability to express, purify and determine their atomic structure, we have learned a great deal about the principles that govern structure and activity of membrane proteins. For example, the ion channels and transporters that mediate the movement of charged molecular species across the lipid bilayer, are now rather well understood; how G-protein coupled receptors (GPCRs) signal to the inside of the cell upon binding to an extracellular ligand is also being elucidated. Indeed, we have acquired a wealth of knowledge about how membrane proteins are shaped and work to allow solutes and biological signals to traverse and be transmitted across the water-impermeable lipid bilayer.
A snapshot of any and every one of such molecules at the atomic level is of invaluable importance in understanding how the membrane protein in question works in both physiological and pathological states of the cell. Indeed, an atomic level structure is unique in providing detailed insight into these cellular processes at a molecular level, revealing the precise protein residues involved in substrate recognition, catalysis or translocation, and, overall, providing us with hypotheses, which can then be tested by functional assays and biophysical techniques.
Membrane proteins constitute 30% of all human proteins and ~60% of drug targets. Yet, they only represent less than 2% of protein structures determined to date. Obtaining a macromolecular structure typically entails measuring X-ray diffraction data from suitable protein crystals or viewing suitably prepared protein samples by single-particle cryo-electron microscopy (cryo-EM). Producing crystals sufficiently ordered to yield quality X-ray diffraction or samples with the appropriate characteristics to allow high-resolution cryo-EM analysis, thus allowing structural investigation, are the key and most problematic steps. This is especially difficult for membrane proteins because they naturally reside in the cellular membrane, and therefore have to be biochemically extracted for purification and structure determination, often leading to destabilization and denaturation.
The Structural Biology Program within the Department of Physiology and Cellular Biophysics strives to determine high-resolution structures of the main protein components involved in cellular events in and across the membrane. The program brings together the expertise of Dr. Wayne A. Hendrickson, Dr. Filippo Mancia and Dr. Andrew Marks within the Department, of Dr. Lawrence Shapiro and Dr. Alexander Sobolevsky from the Department of Biochemistry and Molecular Biophysics, and of Dr. Qing Fan from the Department of Pharmacology.
We have assembled a common core facility for automated set-up, storage, visualization and optimization of crystallization experiments, which is unique and absolutely state of the art, and we interface with the outstanding cryo-EM facilities both at CUMC and at the New York Structural Biology Center (NYSBC). Furthermore, Drs. Hendrickson, Mancia and Shapiro are all active participants of the NIH-funded Center on Membrane Protein Production and Analysis (COMPPÅ; https://www.comppaa.org/), in which at a central facility located in the close-by NYSBC, high-throughput methodologies are used to select membrane proteins with increased probability of yielding structural information.
Qualified graduate students and postdoctoral scientists are encouraged to apply for positions
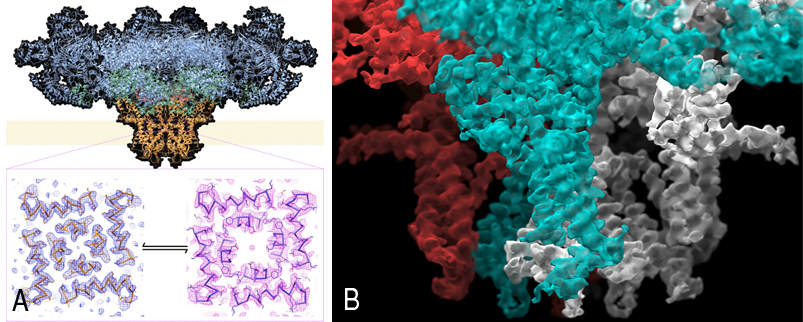
Figure A shows the structure, determined by cryo-EM of the entire mammalian ryanodine receptor and density for the open and closed pores. Figure B shows a close-up image of a cryoelectron microscopy reconstruction of the mammalian ryanodine receptor, focusing on the transmembrane region, with two adjacent subunits colored in contrasting colors.
|
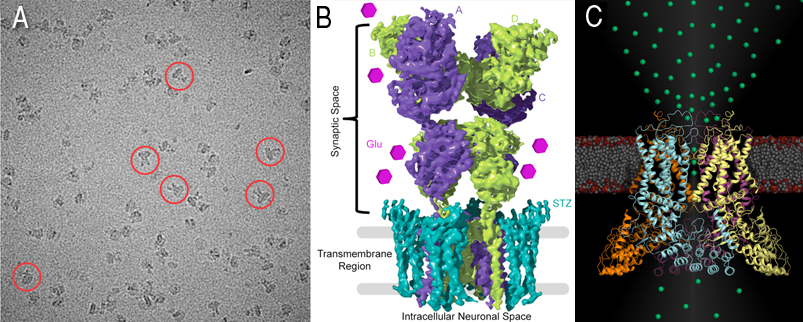
Two images shown above, one for glutamate receptors (Cryo-EM micrograph) with example particles highlighted by red circles (A) and 4.2 Å reconstruction of the GluA2-stargazin complex, with GluA2 subunits colored green and purple, and stargazin in blue (B). Molecules of the neurotransmitter glutamate are shown as pink hexagons. (C) TRPV6, structural representation of the epithelial calcium channel TRPV6. For the first time, calcium ions (green spheres) are crystallographically observed in a calcium selective member of the voltage gated ion channel superfamily, revealing atomic details of calcium selective permeation.
|
|


